Lab Planner
Lab Planner is a Java application created as a final project for BioE 134 (Genetic Design Automation) at UC Berkeley by a group of ten students. It attempts to solve problems inherent in running an undergraduate genetics laboratory, and it was used in this capacity the following semester for BioE 144L (Protein Informatics Laboratory). It can generate steps for an experiment with minimal input, schedule those steps, assign them to students, and automatically update the inventory of the lab as tasks are completed.
For more information on the issues unique to running this sort of laboratory, and how we attempted to address them, see below:
Abstracting Common Design Patterns
Many operations performed in genetics experiments are rather complicated, but the steps performed differ in only a few places depending on the desired product or the starting reactants.
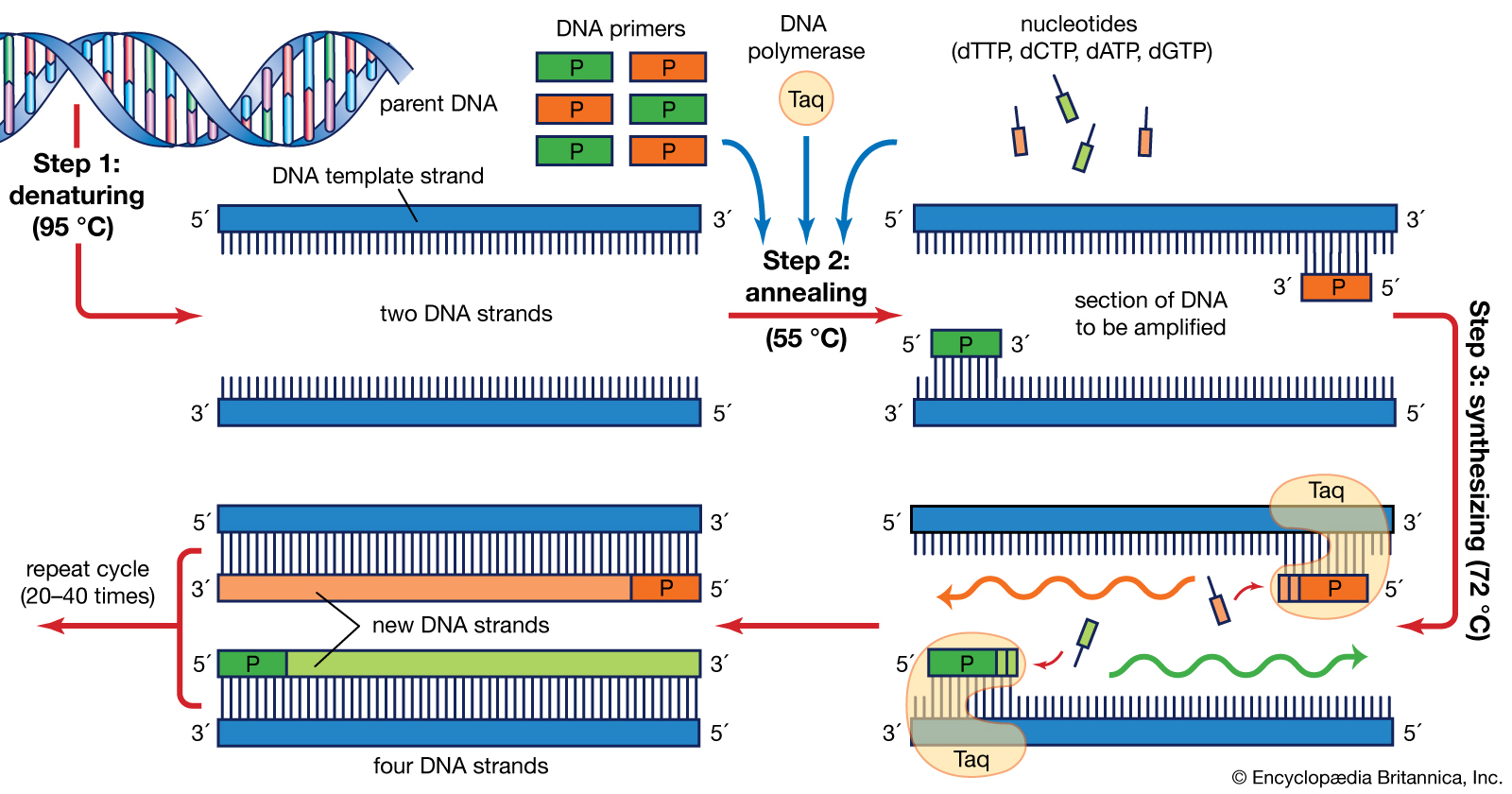
For example, these are steps one might take when performing PCR (Polymerase Chain Reaction) :
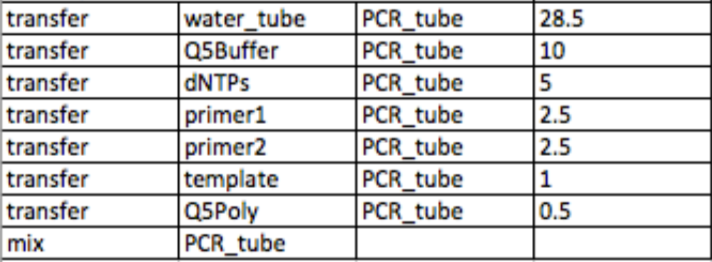
- For each primer oligonucleotide, prepare a 10 micromolar dilution
- Prepare a 20x dilution of the template
- Add 28.5 μL of water, 10 μL of the polymerase buffer, 5 μL of each dNTP, 2.5 μL of each primer solution, 1 μL of the diluted template, and .5 μL of the polymerase enzyme
- Mix well
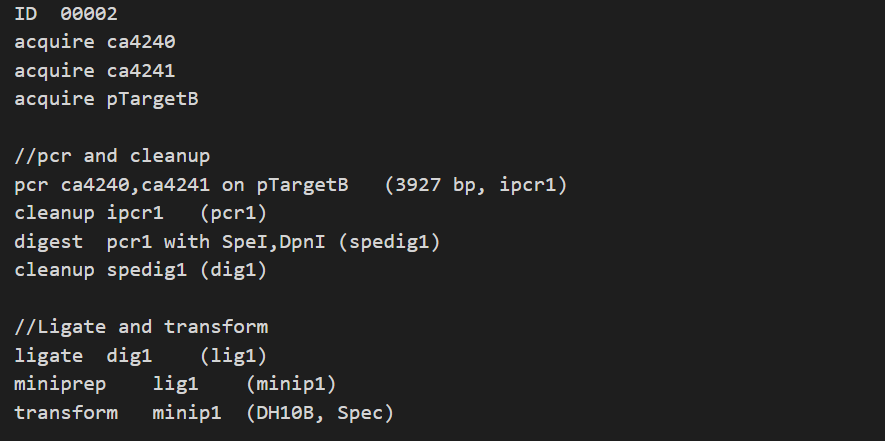
However, the only thing that will change each time you perform PCR are the primers and template. Since a given experiment will have at least a dozen of these operations, it is a lot of redundant information to list out all the steps each time. Therefore, we represent each experiment as a Construction File, where each line of the file condenses an operation as a single step.
For example, the above PCR would simply be represented as pcr [Oligo1],[Oligo2] on [TargetName] ([ProductLength] bp, [ProductName]).
Each step in a Construction File can be assigned to a student, and after they complete it, it will be parsed to update the inventory automatically.
Assigning Asynchronous Tasks to Students
While many introductory-level laboratory courses assign all students the same tasks in a given lab section, in other contexts it is common to divide up the steps for an assignment among students, or even for students to be doing different experiments entirely. If a user to specifies a collection of Construction Files, each representing an experiment, Lab Planner will divide the steps for those experiments among students in a way that minimizes the amount of time students will have to wait in-between steps (e.g. if they have to wait for a shared resource, or if one of the steps requires a certain amount of time to pass before proceeding).
Additionally, if not all steps are completed in a day, they will be rolled over to the next day and Lab Planner will assign steps accordingly. It is even possible to specify absences or holidays, or to only assign students to steps that they have received training for.
Explaining Tasks In Granular Detail
Although a veteran geneticist may know exactly what to do when you tell them something as abstract as “perform Golden Gate Assembly on these fragments using this enzyme”, a student will likely need more information. This might include the exact concentration of solutions they must create, the intermediate steps they must perform, and even what equipment to use. To assist with this, when dividing up steps, Lab Planner will convert each Construction File step to a less abstracted format we call a Semi Protocol.
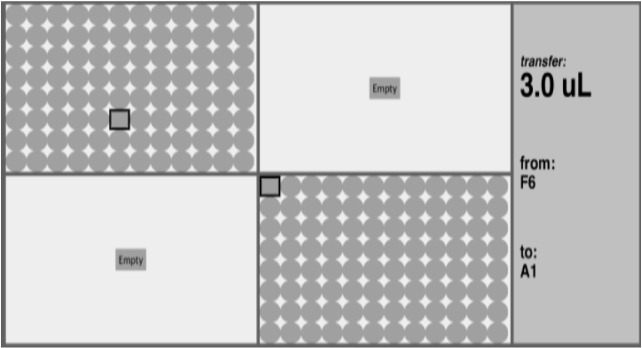
A Semi Protocol describes the steps at a level of granularity appropriate for a student with the prerequisite experience required for an upper-division laboratory course. In fact, a Semi Protocol is sufficient to develop a range of tools to assist with lab procedures. One such application that we considered is a “pipette aid” - a GUI which tells laboratory workers exactly which wells of a microplate to add a liquid to at each step of the experiment.